Bioimage Model Zoo
The Bioimage Model Zoo (bioimage.io) is a recent initiative to make deep learning models for bioimage analysis more accessible and transferable between software.
The Bioimage Model Zoo does two main things:
provides a place to find and download deep learning models relevant for bioimages
standardizes the crucial metadata that accompanies the models
The second relies upon a model spec, stored in a rdf.yaml file.
This includes information such as
where the model weights exist
what inputs are required
what outputs are expected
licensing
authorship
how to cite the model if you use it
It also provides test inputs and outputs, so it’s possible to check the results are correct.
QuPath Bioimage Model Zoo extension
QuPath aims to support the zoo via the QuPath Bioimage Model Zoo extension.
The overall aim is to enable models kept in the Zoo to be imported into some QuPath-friendly form. Currently, the zoo contains a lot of models devoted to image segmentation - so the extension focusses on converting these models to QuPath pixel classifiers.
Adding Deep Java Library
To use the model zoo in QuPath, you’ll need the QuPath Deep Java Library extension.
Not everything works yet!
This is still early and experimental - subject to change in later releases!
Only a subset of models are currently supported, partly because
not all models are Java-friendly, and
some models require accessing all the pixels in the image… but QuPath is designed for huge images, and doesn’t easily support these kinds of global calculations yet
As a result, applying the model in QuPath can sometimes give a different result to applying it in other software. This should improve in future releases.
From zoo model to pixel classifier
To convert a model zoo model to a QuPath pixel classifier, you’ll need:
QuPath, with both the Deep Java Library extension and Bioimage Model Zoo extension
See Deep Java Library for more info about installing & downloading the required deep learning engines
A compatible model from bioimage.io
This is usually in the form of a .zip file, that you’ll need to unzip first
If you download a TensorFlow model, you should also unzip the model weights (saved model bundle)
Some good fortune
With the extensions installed, run the command and select the model specification file.
This is usually called rdf.yaml, although might be model.yaml for some older models.
If all goes well, QuPath will check the model is compatible and show a dialog:
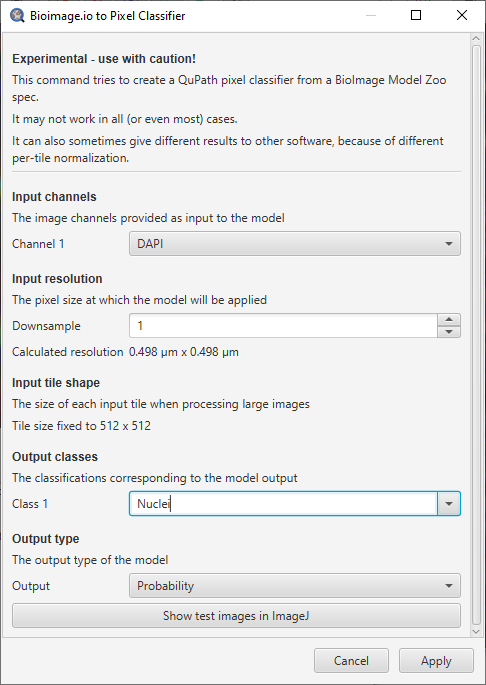
Dialog showing pixel classifier options for a model zoo model.
Figures here were generated using the unet2d_nuclei_broad model spec.
This provides an opportunity to customize a few QuPath-specific aspects:
the input channels (for a multi-channel image)
the input resolution
the input tile size, if this is customizable
how to interpret the output classifications
how the output should be interpreted (generally leaving it at Probability) will be best
There should also be a button to Show test images in ImageJ. This applies the prediction using QuPath to some small sample images included with the model, and compares them with the intended output.
After pressing this button, QuPath will open ImageJ and show the input test image, the target prediction, the actual prediction, and the difference between them.
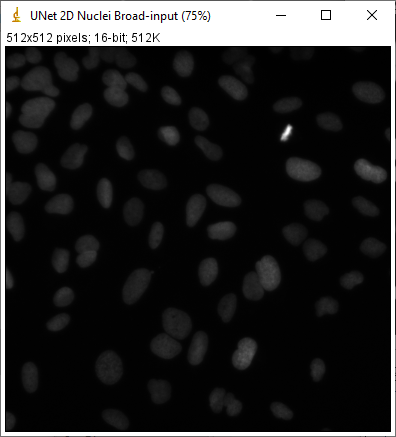



The target and actual predictions should ideally be identical, but can differ - either because of different deep learning libraries, or (more likely) because QuPath is handling tiling and padding in a different way. I hope that any differences will be reduced/eliminated in future releases.
Finally, choosing Apply should give a dialog prompting for a pixel classifier name. Entering this should give a pixel classifier that acts just like any other pixel classifier in QuPath - and so can be used to generate objects, make measurements, or apply classifications.
Which models are compatible?
Model compatibility is a tricky thing. Models in the zoo are currently often compatible with only one or two software applications.
QuPath currently aims to support:
Models that take a single 2D input image and give a single 2D output image (+ channels)
Models without custom pre/post-processing
StarDist uses custom post-processing - so you should check out StarDist instead
Preprocessing with a fixed scale factor and offset is generally fine; preprocessing that requires global statistics (e.g. percentile normalization) may ‘work’, but give different results because these values are calculated per image tile and not across the full image
Models with compatible weights
Assuming you’ve DJL installed, this means:
TensorFlow saved model bundles (you’ll need to unzip the bundle)… assuming you’re not using Apple silicon
PyTorch using Torchscript only
ONNX might work via QuPath’s built-in OpenCV (if you’re very lucky), or if you build QuPath from source adding the OnnxRuntime engine to DJL